Exploiting and sharing existing and new knowledge in the frame of the Digital Botanical Gardens Initiative
Project Coordinator
Pierre-Marie Allard, University of Fribourg, Switzerland
Project Members
- Emmanuel Defossez, Institute of Biology, University of Neuchâtel, Botanical garden of Neuchâtel, Switzerland
- Adriano Rutz, Institute of Molecular Systems Biology, ETH Zürich, Switzerland
- Ana Claudia Sima, SIB Swiss Institute of Bioinformatics, Switzerland
- Tobias Kuhn, Knowledge Pixels, Switzerland / VU University Amsterdam, Netherlands
- Tarcisio Mendes de Farias, SIB Swiss Institute of Bioinformatics, Switzerland
BiCIKL Contact person
Patrick Ruch, SIB
BiCIKL Research Infrastructures involved
SIBiLS, Plazi, EBI (ChEBI), CERN (BLR, Zenodo), OpenBioDiv
Non-BiCIKL Research Infrastructures accessed
NCBI (PubChem), Wikimedia foundation (Wikidata), Nanodash (Nanopublications)
Biodiversity data classes and services included
- Plants
- Metabolites
- Open Access and paywalled literature
- Biodiversity
- Chemodiversity
- Open Science
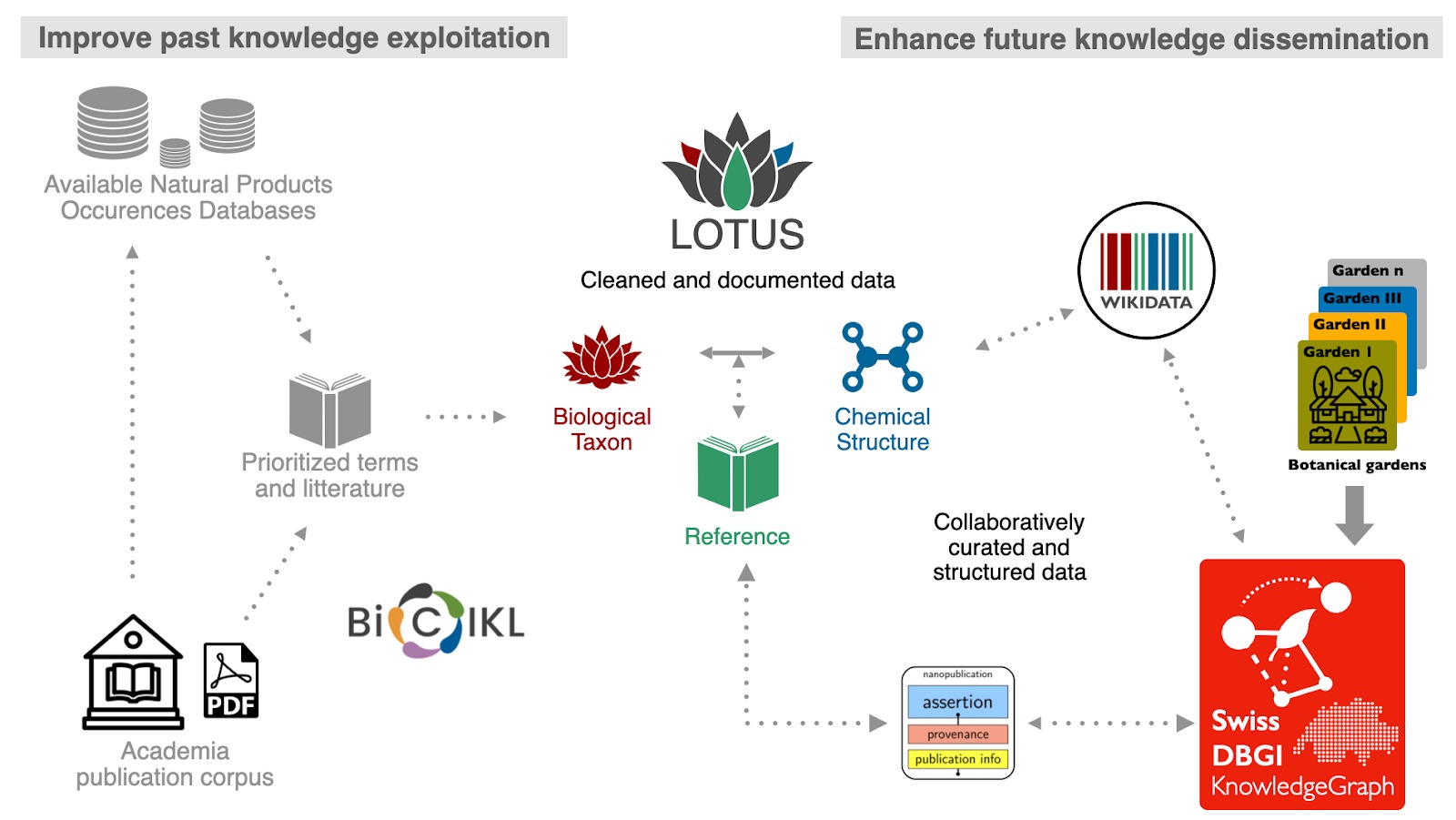
Fig.1. Improving the exploitation of publicly available past knowledge and building news forms of knowledge dissemination for natural products research.
Background
We are currently launching the Digital Botanical Gardens Initiative (DBGI) with the ambition to explore innovative solutions for the acquisition, management and sharing of digital information acquired on living botanical collections. A particular focus is placed on the large-scale characterization of the chemodiversity of living plant collections through mass spectrometry. The acquired data will be structured, organised and connected with relevant metadata through semantic web technology. After validation and application in wild ecosystems, the gathered knowledge will inform ecosystem functioning research and orient biodiversity conservation projects.
One central aspect of the DBGI thus resides in the acquisition of high quality information on the digitised chemodiversity which in turns rely essentially in efficient metabolite annotation. For this we employ a taxonomically informed metabolite annotation process (https://doi.org/10.3389/fpls.2019.01329), which we have shown to systematically improve the performance of state-of-the computational metabolite annotation solutions. A requirement for this process is to access comprehensive ressources documenting the biological occurrence of small molecules. For this we recently opened the LOTUS ressource (https://elifesciences.org/articles/70780) to the community. However, numerous biological occurrences of natural products are still lacking.
We would like to evaluate the BiCICKL Research Infrastructures to:
- Improve past knowledge exploitation: extract pairs of chemical compound and taxon from literature not present in LOTUS and relevant to the DBGI (papers describing phytochemical investigation of plants of the Swiss Botanical Gardens, e.g. https://doi.org/10.1016/j.phytochem.2020.112469)
- Enhance future knowledge: explore publication solutions to efficiently disseminate knowledge acquired from metabolomics, that is, putative occurrences of chemical structures in biological matrices (simple RDF triples, Nanopublications or similar).
Expected outcomes
Through the combined expertise of SIBILS, ARPHA, Plazi, BLR, GBIF and OpenBioDiv we expect to achieve the following outcomes.
For the “Improve past knowledge exploitation” part:
- A tabular file compiling referenced chemical structure - organism pairs extracted from literature. Should contain SMILES, IUPAC and/or common name of chemical structures; binomial denomination of organisms; and DOI of the reference documenting this occurrence. Additional metadata (material citation, collections, treatments, and specimen information) will complement the occurrence description. Will be contributed to LOTUS.
For the “Enhance future knowledge dissemination” part:
- A nanopublication (or alternative format) reporting the occurrence of metabolites evidenced in a computational mass spectrometry experiment.
Results
During this project, 1,742 PDF files totalling 17,084 pages from the Phytochemistry journal have been processed through the Plazi pipeline and 554 taxonomic treatments, 8,613 figures and 532 tables have been extracted and shared. Several millions of LOTUS terms have been highlighted through the SIBILS pipeline and can now be observed at https://sibils.text-analytics.ch. A total of one million annotations (N=99 905 210 on March 2024) have been generated into SIBiLS. Each has been associated with a bi-directional link to a WikiData page for a given LOTUS compound (see example in Figure 4.5 below). For each natural chemical compound, a unique identifier was assigned and a BioC micro-citation instance was created in the SIBiLS article record. Figure 4.6 shows the landing page for the compound in WikiData. This project is led by the BiCIKL partners SIBiLS and Plazi and performed by a research group from Switzerland led by Dr. Pierre-Marie Allard from the University of Fribourg.