Interactions of taxa based on public DNA sequences in INSDC (NCBI, ENA, DDBJ) databases
Project Coordinators
Urmas Kõljalg, Kessy Abarenkov, Allan Zirk
Project Members
Yui Fujinuma, Henrik Nilsson, University of Gothenburg; Julia Pawlowska, University of Warsaw
BiCIKL Research Infrastructures involved
PlutoF, ENA, GBIF, DiSSCo / Naturalis, TreatmentBank, CoL, ARPHA-XML
Non-BiCIKL Research Infrastructures accessed
NCBI (GenBank)
Biodiversity data classes and services included
Sequences, specimens, living specimens, material samples, taxon names, literature;
Services: annotation datasets are available for download in different formats (FASTA files used by different analytical pipelines like QIIME, mothur, CREST, etc. CSV files downloaded from PlutoF).
Background
This project explored the functionality provided by the PlutoF platform to identify, connect and store biodiversity data, with a starting point of DNA sequences derived from various experimental approaches using fungi and higher plants. As is typically the case, sequences obtained through DNA sequencing approaches can be poorly annotated across various databases, hindering data interpretation. This project explored approaches to improve those annotations, overlaying taxon name, source material, taxon interactions, geolocation data, synonyms, and other data to provide a more holistic knowledge set around each species.
Expected outcomes
The expected outcome was an improved annotation of 200-500 sequences to assist in interpretation of other experimental data.
Methods
With a starting point of a set of experimentally derived fungal and plant sequences, Yui Fujinuma systematically reviewed and linked data between ENA, GBIF, and specimen collection records using PlutoF platform to persistently store the new linkages.
It involved reviewing literature where INSD sequence records of Japanese plants and fungi were originally published. As part of the annotation work, culture (Figure 2) and specimen records (Figure 2) were retrieved from an assortment of collections and linked to the DNA sequences (Figure 3). Specimen and sequence records were annotated with taxon names (which are often missing or too ambiguous in public sequence databases), and taxon names missing from PlutoF classification were imported from GBIF backbone taxonomy. Improved taxon identifications and annotations to sample source and geolocation were sent to ENA via ELIXIR Contextual Data ClearingHouse API - these were linked to the corresponding sequence record and are available for public use.
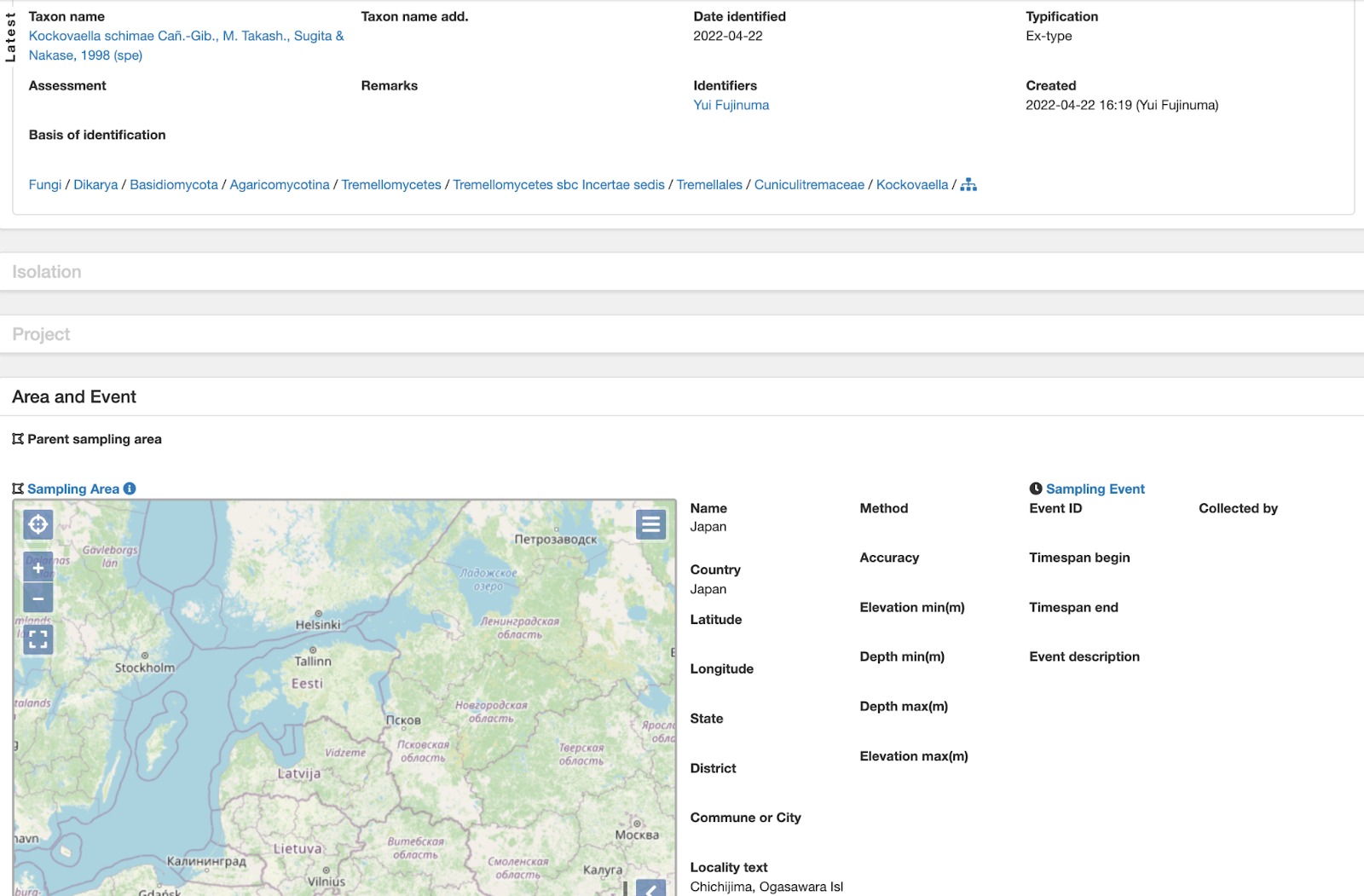
Fig. 1. Showing living culture record added to PlutoF during the annotation process.
Results and Discussion
During this small project, 380 INSD sequences were annotated) source metadata and geolocation data was improved and made available to ENA, b) new linkages were made across ENA and GBIF by linking INSD sequence records to collection specimens and living cultures for which digital records are present in GBIF and/or other collection databases.
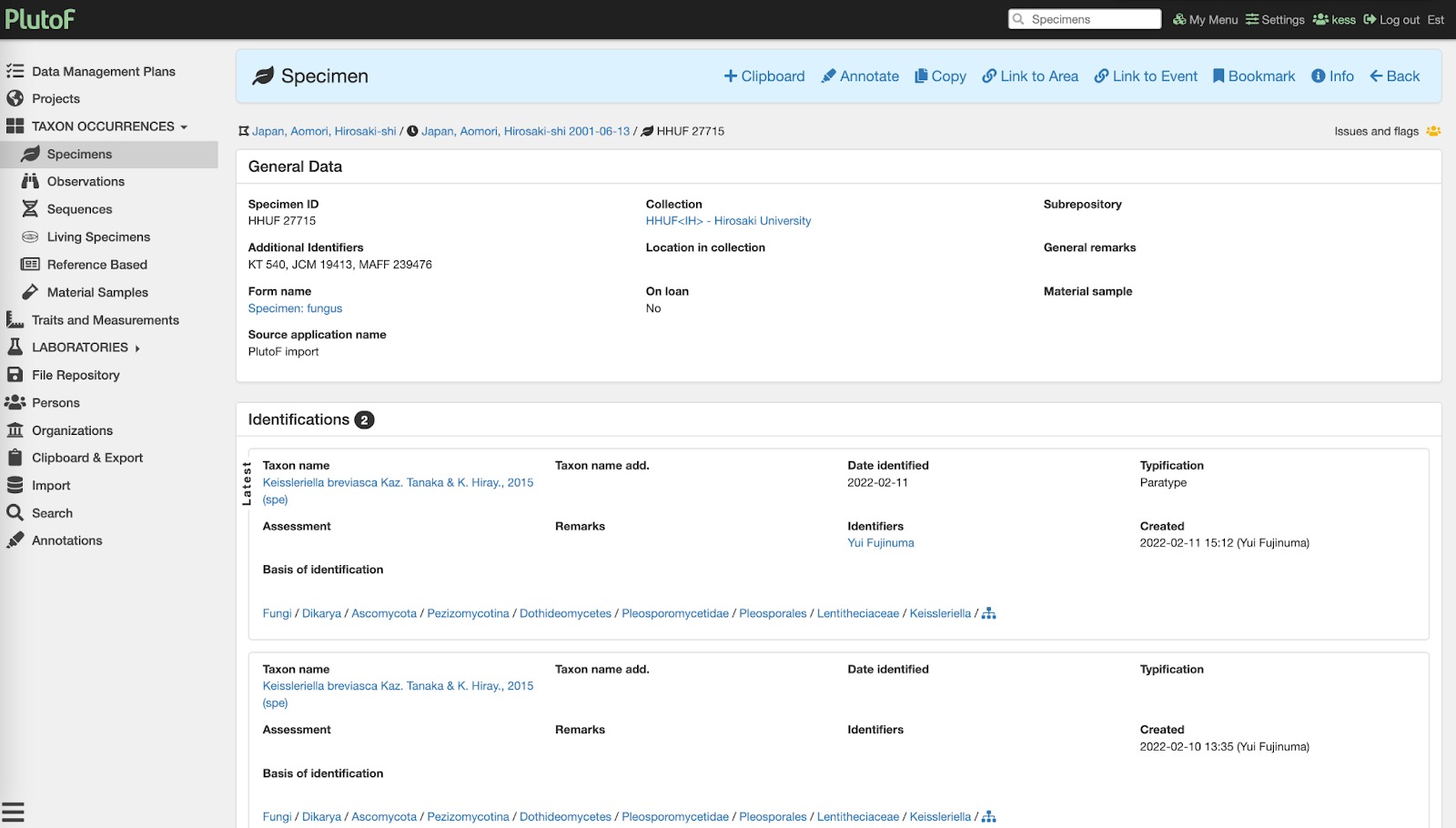
Fig. 2. Showing specimen record added to PlutoF during the annotation process.
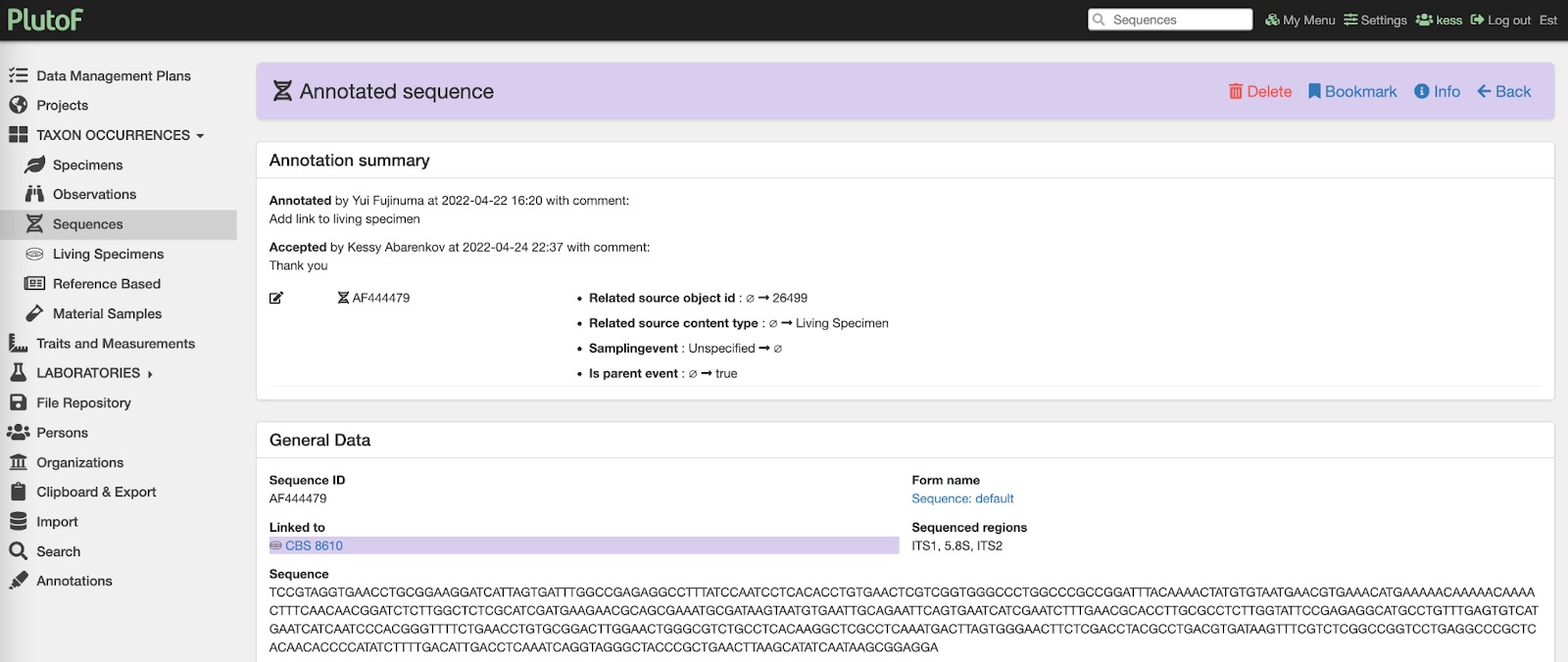
Fig. 3. Showing example annotation summary from the PlutoF platform where INSD sequence is linked to specimen record presented in Fig. 2.
Future Work
There is likely to be opportunity for further enhanced annotation of the data set that was explored in this pilot study, especially as new data is likely to emerge across many of the data sets under consideration. In addition, it has proved a valuable exercise confirming the utility of PlutoF as a useful hub for Biodiversity related sequence data. It would be interesting to explore other data sets and applications via the BiCIKL Open Call process, that have a focus on PlutoF are very welcome.