Lestid Damselflies: linking digital data sets, treatment materials citations, collections databases, and phylogenies, to enhance data exchange
Project Coordinators
Jeremy Miller, Naturalis and Donat Agosti, Plazi (Lead)
Project Members
Jeremy Miller, Naturalis; Hector Ortega Salas, Naturalis; Alperen Bektas, Berner Fachhoschule; Klaas-Douwe B. Dijkstra, Naturalis
BiCIKL Research Infrastructures involved
TreatmentBank, BLR, Zenodo, CoL, GBIF
Non-BiCIKL Research Infrastructures accessed
BioPortal, BOLD, NCBI
Biodiversity data classes and services included
Specimens, Literature, Taxonomic names, specimen digitization, DNA sequences
Background
The lestids, known commonly as the spreadwings, are a cosmopolitan family of large-sized damselflies of 150 species (CoL). To study this taxon, ongoing work aims at understanding how well they have spatio-temporally been collected, where major lacunas occur and thus where collecting efforts should be guided.
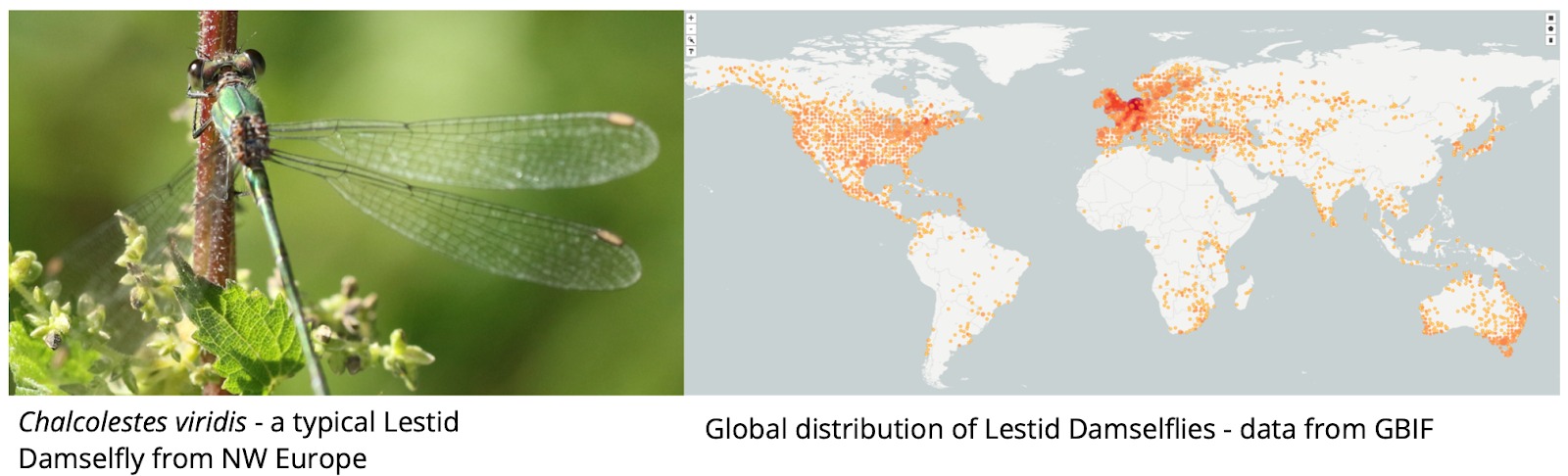
Fig. 1. Typical data available for an example Lestid Damselfly.
The taxon is well represented in Naturalis covering specimens of many species, a major part of the literature has been digitised and added to the Biodiversity Literature Repository, and DNA sequences have been accessed by online databases.
The goal of this project was to create a cyber catalogue by linking treatments via treatment citations to cited, previous treatments, link material citations in taxonomic literature, specimens in natural history collections databases, and DNA sequences in scientific literature and databases, using persistent unique identifiers (PIDs), and to develop bidirectional links to enhance database records.
The results will be part of the cyber-catalogue of this taxon, with linked entries in BLR, GBIF, Naturalis, ENA (?) and COL+. This is an ideal basis to develop bidirectional linking and explore how the data is best provided to the respective research group at Naturalis. We intend to make annotations available to collections databases when a specimen in their collection has a DNA sequence in GenBank, or when a taxonomic name changes. We also fill in missing data fields in a collections database from treatment material citations, or flag discrepancies. Figures cited in treatments and images of collections material are also potential for sharing.
Expected outcomes
We envisaged that researchers will gain access to species occurrence records currently unavailable in digital form. This data will contribute to assessments of species distribution including more accurate species distribution models, more accurate information from which to plan field work for recollecting known species, and baseline information for better assessing changes that may be attributed to climate change.
We extracted primary taxa on the literature on the Lestid family of damselflies, in phases, starting with the “low-hanging” fruit, papers, the ones that had material, citations records, the ones that were more important. We marked these up, using the Plazi infrastructure and them to contextualise this information with GBIF.
We were aiming to annotate all of our literature records for which we could find the same specimen in the museum collection database with identifiers for those museum collections. The second phase was to build a catalogue, basically an index of all original descriptions of all the species, their important changes, the different names they may have been known as for the last 200 years.
Results and Discussion
The work carried out in this short project has resulted in a significant enhancement in the data around this family of damselflies. 234 treatments extracted from the literature using the Plazi infrastructure are now available in Treatmentbank, of which 210 treatments across 130 species and 246 material citations have been added to the GBIF database.
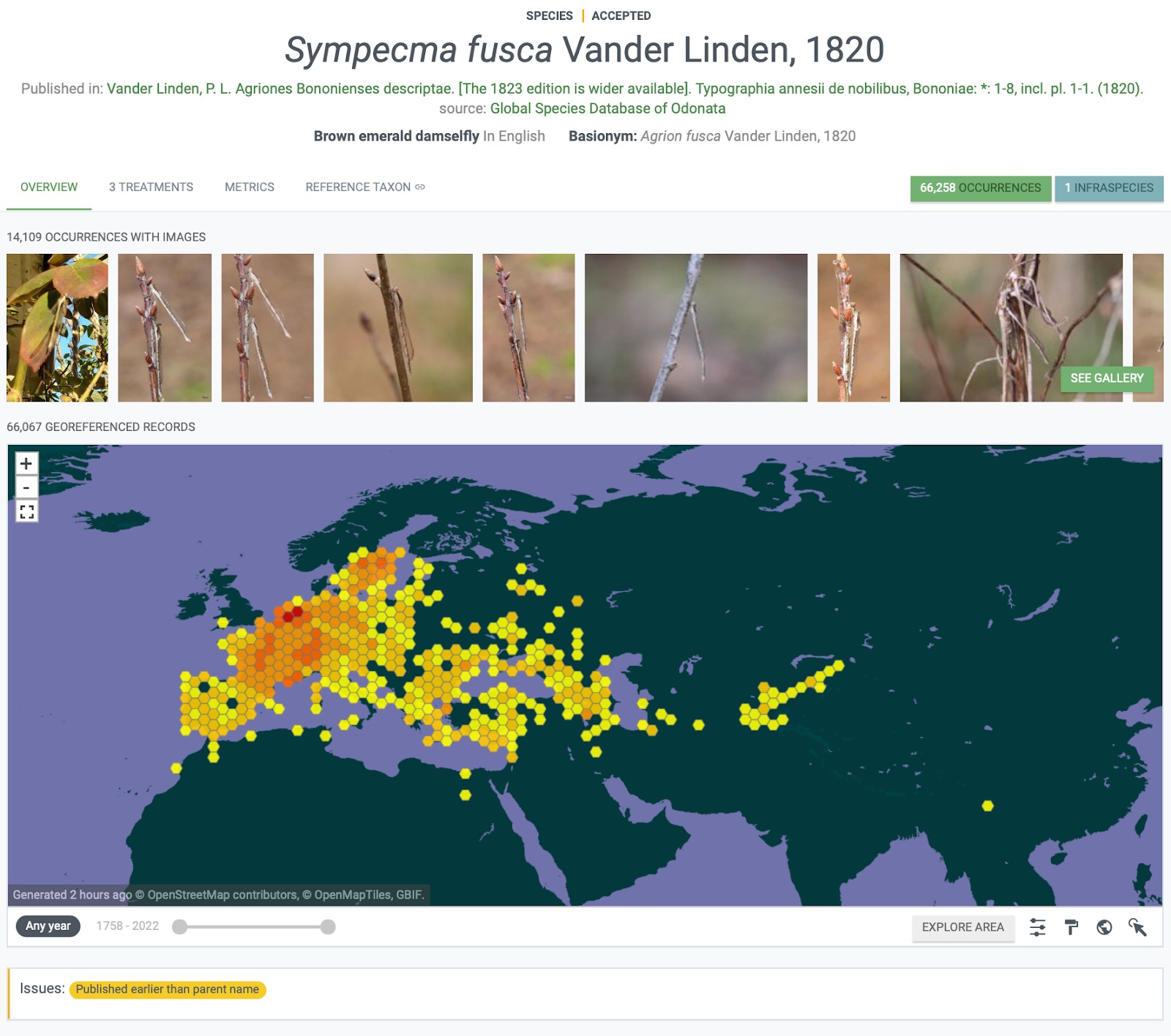
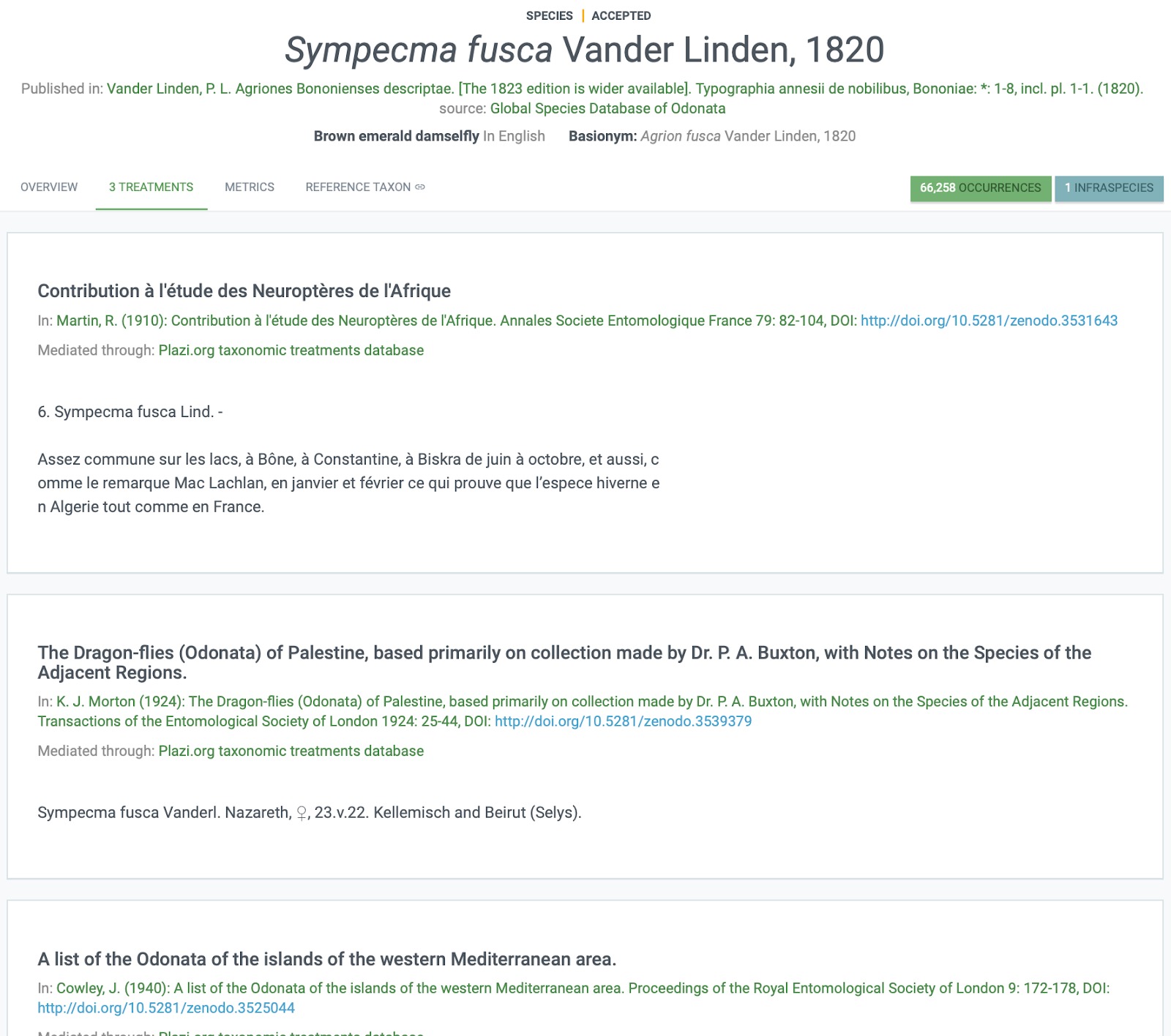
Fig. 2. Illustrating that 3 new treatments were added to the Lestid Damselfly, Sympecma fusca.
This has significantly enhanced the taxonomy and understanding of the species in this group, and has made significant progress in the longer term aim to build a very high quality species catalogue for Lestid damselflies, including not only species name but also all known synonyms, treatments and from GBIF observational data the distribution of each species.
Future Work
This project has built a foundation for the establishment of a high quality curated species catalogue for Lestid Damselflies. This of course can continue to be built upon and refined as further data becomes available. Furthermore, it has established workflows and techniques that could be used in other taxa groups, to work in a similar fashion, and it would be interesting to extend this work to other groups.