New tools to highlight and shortlist enigmatic fungi from eDNA datasets for further scientific exploration
Project Coordinator
Henrik Nilsson, University of Gothenburg, Sweden
Project Members
Henrik Nilsson, University of Gothenburg, Sweden; Julia Pawlowska, University of Warsaw, Poland
BiCIKL Contact person
Urmas Kõljalg, UTARTU
BiCIKL Research Infrastructures involved
PlutoF, CoL, GBIF, ELIXIR and ENA
Non-BiCIKL Research Infrastructures accessed
UNITE, MycoBank, IF
Biodiversity data classes and services included
DNA sequences, taxon names, specimens, living specimens, curating (third-party annotations) of DNA sequences
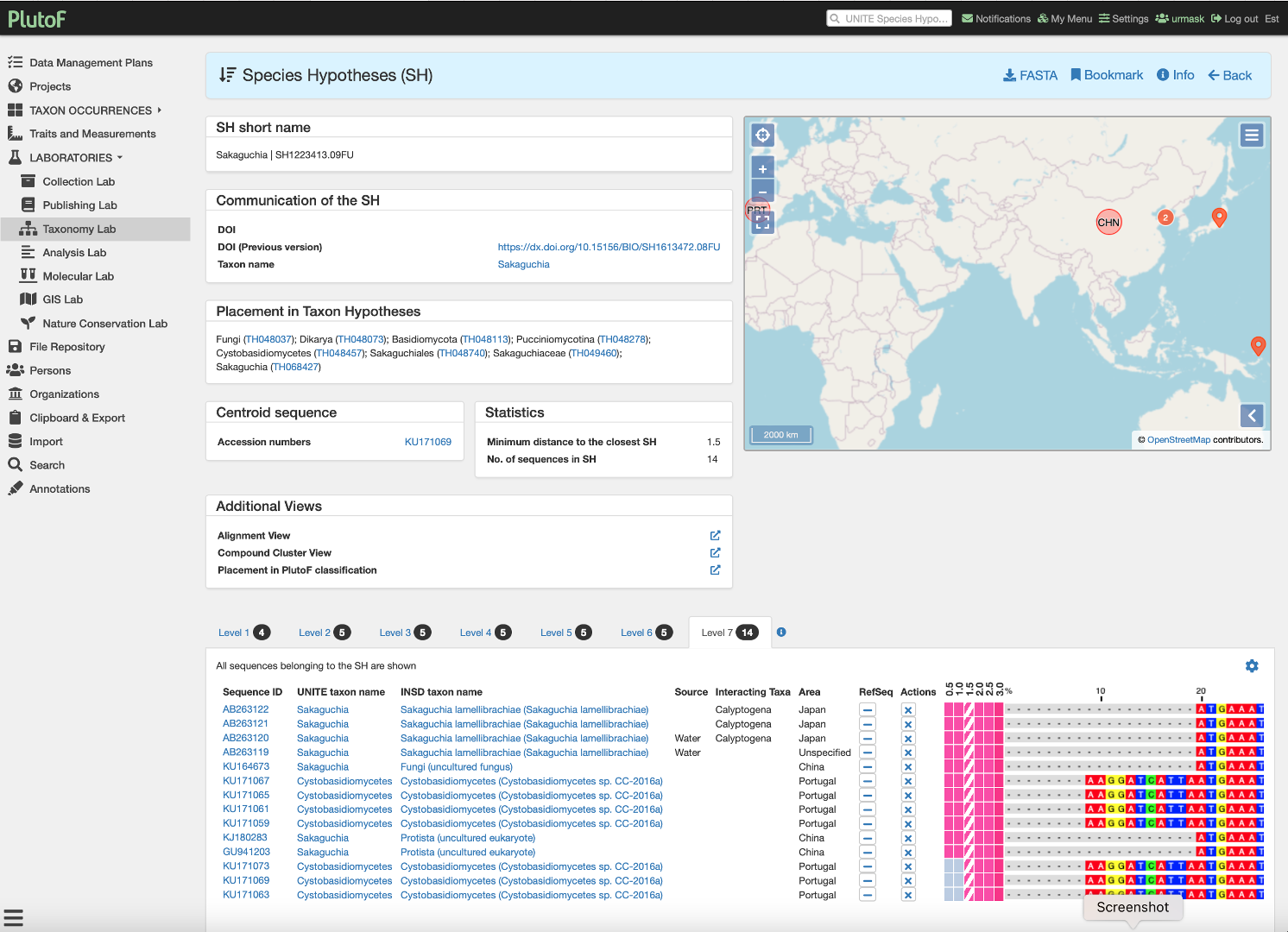 Fig.1. Screenshot of the single UNITE Species Hypotheses in PlutoF. Here researchers can curate eukaryote species based on public rDNA ITS sequences. Curated third-party annotations can be sent directly to the European Nucleotide Archive and they are included in the UNITE reference datasets used in eDNA analyses.
Fig.1. Screenshot of the single UNITE Species Hypotheses in PlutoF. Here researchers can curate eukaryote species based on public rDNA ITS sequences. Curated third-party annotations can be sent directly to the European Nucleotide Archive and they are included in the UNITE reference datasets used in eDNA analyses.
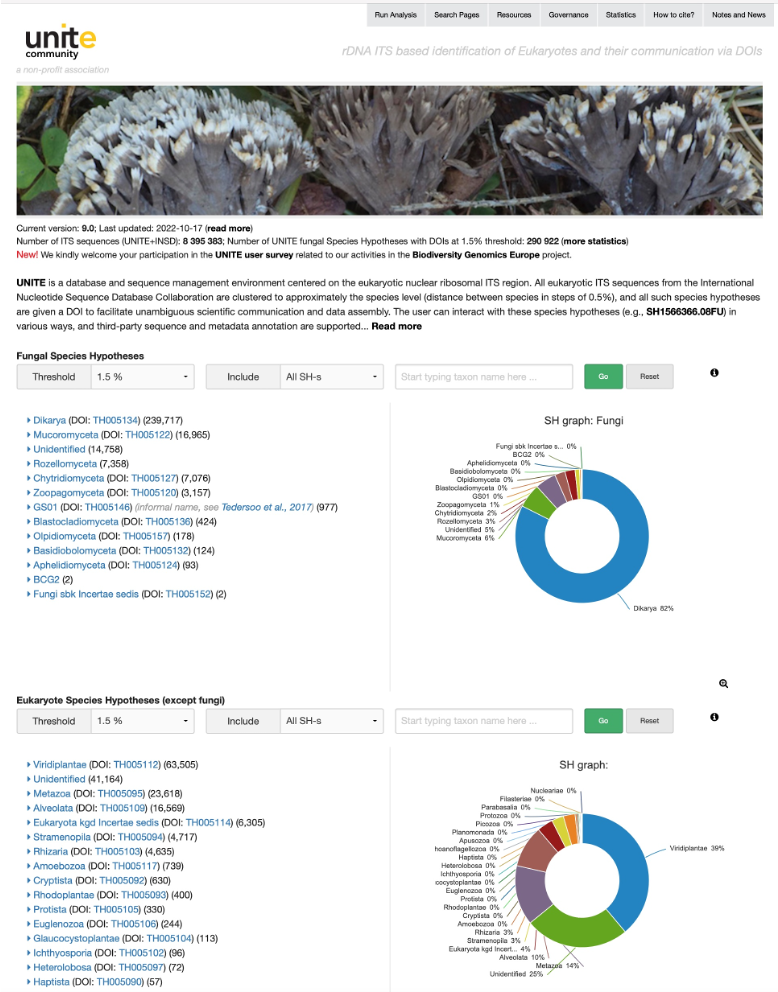 Fig.2. Screenshot of the UNITE homepage displaying the number of fungal and other eukaryotic species hypotheses (SH) in the current version 9.0. Reference databases of SHs can be downloaded from the Resources page in different formats and implemented in different eDNA pipelines like QIIME, mothur, CREST, etc.
Fig.2. Screenshot of the UNITE homepage displaying the number of fungal and other eukaryotic species hypotheses (SH) in the current version 9.0. Reference databases of SHs can be downloaded from the Resources page in different formats and implemented in different eDNA pipelines like QIIME, mothur, CREST, etc.
Background
The fungal kingdom is being redefined by the staggering numbers of hitherto undescribed species unravelled by environmental sequencing. Mycologists have precious few tools to explore these data, leaving the data woefully unexplored for fungal diversity. We propose a collaboration between PlutoF, Index Fungorum (IF), MycoBank (MB), Catalogue of Life (CoL), GBIF, ELIXIR, and European Nucleotide Archive (ENA) to develop a set of software tools to allow fungal taxonomists and ecologists to make use of these data in their research.
- We want at least the best ENA (effectively all INSDC) fungal metabarcoding datasets to be continually incorporated into PlutoF such that they can be explored through the powerful tools offered by PlutoF. The process by which sequence datasets is added is at present semi-automated at best, and its very time-consuming nature essentially prevents addition of new datasets. Having this process automated would be invaluable in the pursuit of mycological and biological progress.
- We’d like a tool that highlights potentially new and undescribed species to speed up their formal discovery and description by the mycological community. This tool would draw from the existing PlutoF/UNITE framework and would, for any genus (or any taxonomic rank of the user’s choosing), highlight all un-named operational taxonomic units (at selected similarity level) that are more than 3% dissimilar from a reference/type-derived sequence, and visualise them with all available metadata and various export functions.
- We’d like to be able to subscribe by email to the taxonomic affiliation of any full-length eukaryotic ITS (barcode) sequence (from environmental and taxonomic sources alike) in PlutoF/ENA as these undergo progressive refinement by incorporation of new taxonomic reference sequences over time. This tool would automate the arduous process of monitoring select INSDC accession numbers for taxonomic affiliation over time, a task that many mycologists do manually (and thus irregularly) right now.
- We’d like new taxonomic names from MB and IF to be added automatically to PlutoF and integrated into the above functions. Roughly 1,500 new species of fungi are described each year, and herbarium sequencing efforts are similarly producing DNA reference sequences for species described decades and sometimes centuries ago. Similarly, mycology sees a flurry of taxonomic and nomenclatural rearrangements in the wake of molecular data. PlutoF currently has a semi-automated nomenclatural engine that cannot keep track of all new names and rearrangements. Thus, a great deal of time is spent by PlutoF/UNITE users to add names and taxonomic rearrangements by hand.
Expected outcomes
Many fungal taxonomists do not have the bioinformatics background needed to extract the taxonomic data from the enormous pool of metabarcoding datasets available, but the tools presented in this proposal essentially get rid of the need for a bioinformatics background to explore eDNA sequences for taxonomic signal. The proposal seeks to blend fungal taxonomy and molecular ecology so that it will be straightforward to draw from molecular ecology results in taxonomy, and the other way around. This is not the case today, and the two fields are often pursued as more or less decoupled fields (https://mycokeys.pensoft.net/article/76053/ and https://mycokeys.pensoft.net/article/56691/).
- Tools 1-2 make it easy for fungal taxonomists to harvest the taxonomic data that is now hiding un-explored in the ocean of environmental sequencing data. The tool removes the technical obstacles involved in locating and targeting potentially undescribed species for further exploration and, in many cases, formal description.
- Tools 3-4 automate a technically complicated and time-consuming process: staying abreast of the taxonomic annotation of sequences and operational taxonomic units of a user’s particular interest. Through subscription, the user would get instant news as new explanatory taxonomic information becomes available, a process that often takes years today.
Results
The project was supervised by the BiCIKL partner UTARTU. Dr. R. Henrik Nilsson (University of Gothenburg) and Dr. Julia Pawlowska (University of Warsaw) lead the research team who used the new services. Many new services and tools were developed or upgraded and deployed in the PlutoF platform for molecular identification of eukaryotes. Examples include importing new taxon names from other infrastructures (Mycobank and GBIF), third-party annotations of INSDC sequences and publishing them in ENA, an SH-matching analysis tool, and publishing eDNA data in GBIF. 8,395.383 HTS (High Throughput Sequencing) and Sanger ITS sequences were linked to the taxonomic backbones of PlutoF and GBIF which allowed these sequences to be published in the GBIF database. More importantly eDNA based studies can now publish their ITS based findings as taxon occurrences in GBIF. Users of the sequence third-party annotation tool made 27,941 annotations from which 4,584 were exported to ENA through Elixir Contextual Data Clearinghouse.