Third-party annotations are a valuable resource to improve the quality of public DNA sequences. For example, sequences in International Nucleotide Sequence Databases Collaboration (INSDC) often lack important features like taxon interactions, species level identification, linkages to source material in scientific collections, information associated with habitat, locality, country, coordinates, etc.
The ambition of Task 8.3 in BiCIKL is to build a web interface for reporting of errors and gaps in sequenced material source annotations by feeding this information back to primary repositories (INSDC).
The ELIXIR Contextual Data ClearingHouse (CDCH) offers a light and simple RESTful Application Programming Interface (API) to report such annotations, either in a single batch operation or on a case-by-case basis, as individual annotations emerge. The data repositories (such as European Nucleotide Archive) then access the staged annotations and enact updates as appropriate for their operations.
The web interface will be provided by PlutoF, an online data management platform for biology and related disciplines that features an annotation module, where third-party annotations can be added to any collection specimen, living culture or DNA sequence record.
There are three stages for conducting the work under Task 8.3 of BiCIKL, of which stage 1 has already been completed:
- Stage 1: Implement third-party annotation submission workflow from PlutoF workbench to the ELIXIR CDCH.
- Stage 2: Implement the user-initiated process of fetching INSD sequence data to be incorporated into PlutoF for annotating purposes.
- Stage 3: Provide public services for retrieving ELIXIR CDCH third-party annotations through community specific portals.
To achieve our specific purpose in Stage 1, making all INSDC sequence annotations added through PlutoF available through the ELIXIR CDCH, we:
- mapped PlutoF and ENA fields available for annotating,
- set up and verified PlutoF annotation workflow by going through a set of third-party annotation use cases,
- created user manual for adding third-party annotations in PlutoF,
- registered PlutoF platform as a service pushing the annotations to ELIXIR CDCH,
- implemented the technical solution in PlutoF.
Prototype for posting annotations to the ELIXIR CDCH test server is available at PlutoF development server.
In the graph below, we illustrate the annotation workflow:
- User annotates sequence metadata.
- An Annotation Proposal is created, and verification notification sent out to the designated reviewer.
- The reviewer either accepts the Annotation Proposal, or rejects it with a comment.
- If the Annotation Proposal is accepted, annotated fields that could be mapped to INSD fields are pushed to the ELIXIR CDCH using their RESTful API.
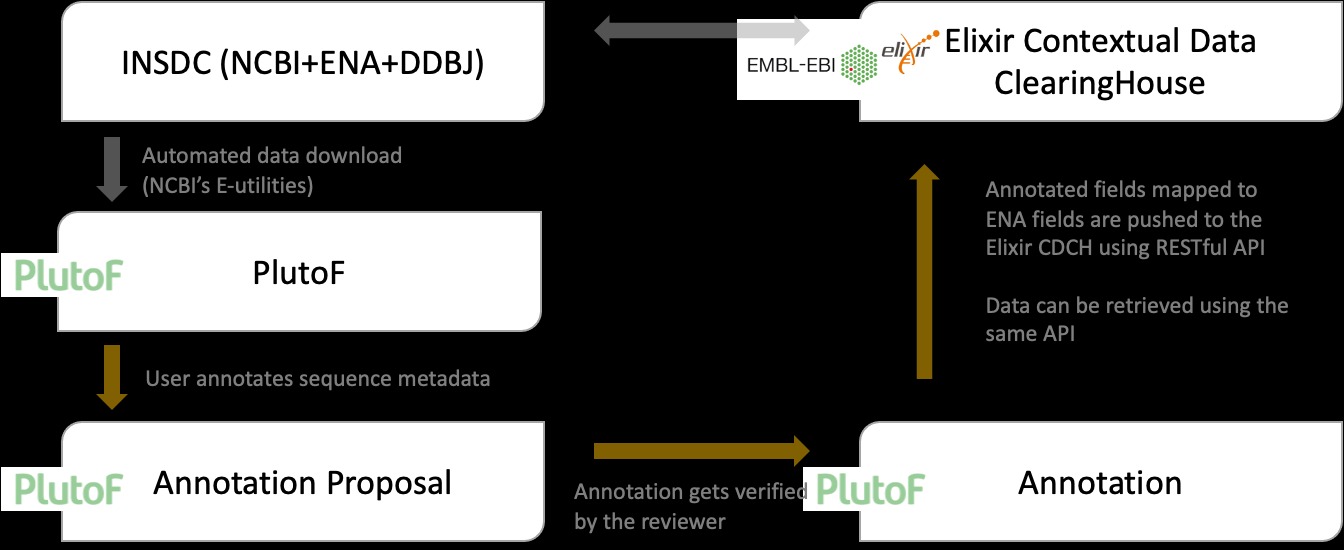
Graph describing how third-party annotations are added and verified in PlutoF and sent to the ELIXIR CDCH.
The results of the work at Task 8.3 in BiCIKL were presented at the 2021 TDWG conference. The conference abstract is openly available in the Biodiversity Information Science and Standards journal (BISS Journal).
