It’s a pleasure today to meet with Pierre-Marie Allard, from COMMONS Lab - Department of Biology of the University of Fribourg, in Switzerland. Allard and his group have proposed a project about “Exploiting and sharing existing and new knowledge in the frame of the Digital Botanical Gardens Initiative”. Let’s see what this is about.
How did you get to know BiCIKL and this call?
We have been following the work of Donat Agosti (Plazi) for some time with a lot of interest and have been aware of the BiCICKL project mostly through the internet and social media.
Why did you apply here, to address what challenge/need?
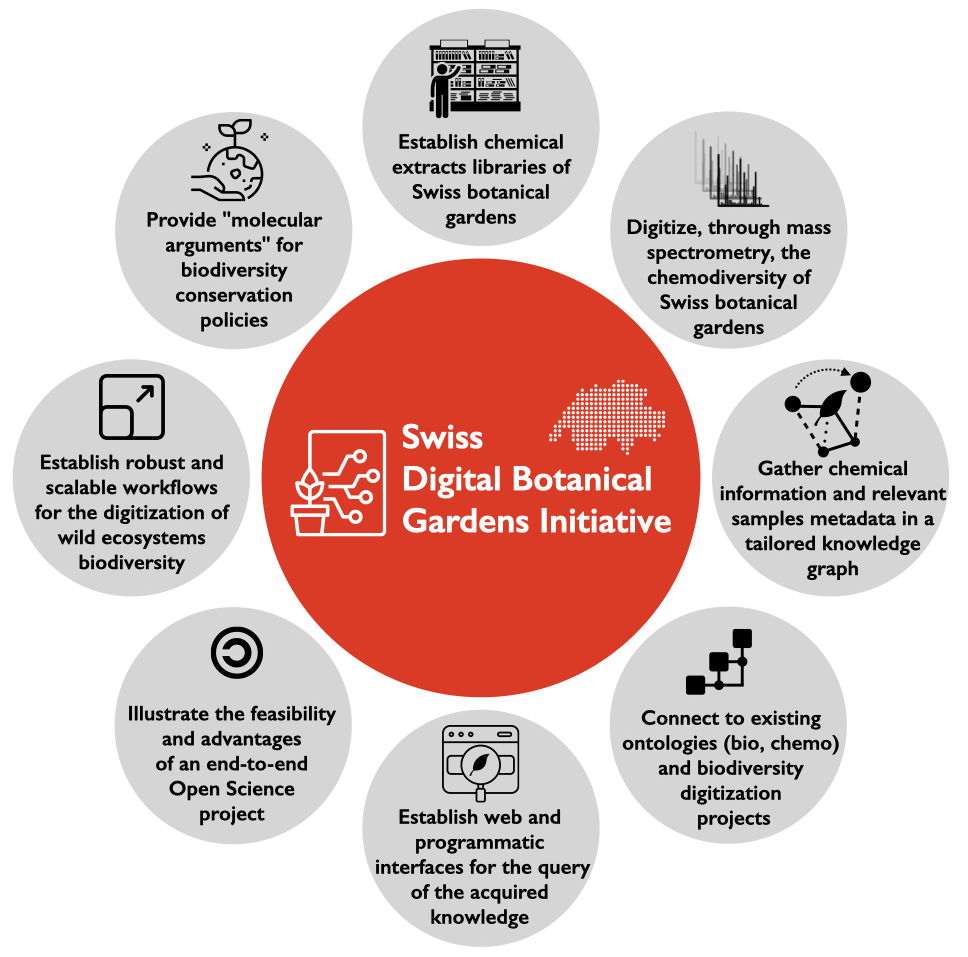
In the last years, we established LOTUS, an open electronic resource documenting the biological occurrences of natural products (https://doi.org/10.7554/eLife.70780). More recently, we launched the Digital Botanical Gardens Initiative (DBGI https://www.dbgi.org/). This Open Science project aims to establish workflows for the characterization of global chemodiversity. In this context, we describe novel chemical information (structures, occurrences, biological activities etc.). For this we depend on access to previous knowledge of natural products chemistry. If some are shared in LOTUS, much of it remains locked in the existing corpus of publications. The extraction of this existing knowledge is the big challenge to address.
How BiCIKL would help to address your need or challenge?
In the frame of BiCICKL, the Swiss Institute of Bioinformatics Literature Services (SIBiLS) and Plazi's TreatmentBank service teamed to propose eBioDiv (https://ebiodiv.org/) a service for Swiss biodiversity scientists to access and disseminate their research data about species in legacy and prospective publications. Building on this experience, we will complement the pre-existing toolbox to mine the literature and extract natural products occurrences related information (chemical structures, biological organisms and their links). These objects will be associated with relevant identifiers and will then be shared in the Wikidata knowledge base where LOTUS lives.
Who will benefit from your project?
As daily users of the LOTUS resource, we will evidently benefit from the expected increase in available information volume. However, the wide availability of the LOTUS resource (through Wikidata or other resources such as PubChem) ensures that this increase in information volume will benefit any researcher interested in the chemical description of living systems.
What will you consider a success for your project?
The establishment of a robust toolbox allowing the automated extraction and sharing of natural products from the existing literature corpus would be, in itself, a success. Obviously, the associated improvement expected in terms of knowledge dissemination is also very much looked after! On a broader perspective, we also expect that such a project will raise the attention of researchers and publishers, on possible best practices to be implemented when reporting research outcomes in the field of natural products chemistry (e.g. tabular files of SMILES, InChI and InChIKey encoded structures together with binomial denomination of taxa).
Please write here a personal quote that resumes the project
Mining historical pharmacognosy and natural products chemistry literature for the future of chemodiversity description.