Linking type specimens to the names they typify for the plant genus Solanum
Project Coordinators
Maarten Trekels and Quentin Groom
Project Members
Roger Hyam, Royal Botanic Garden Edinburgh; Heimo Rainer, Natural History Museum, Vienna
BiCIKL Research Infrastructures involved
TreatmentBank, GBIF, MBG, Naturalis, BGBM, CoL (Species 2000)
Non-BiCIKL Research Infrastructures accessed
Wikidata, IPNI
Biodiversity data classes and services included
Specimens, Literature, Taxonomic names, Text mining, specimen digitization
Background
One of the first jobs a taxonomist must do when revising a groupis to find and examine the nomenclatural type material for the taxa. Nomenclatural types, together with the formal protologue, link a name to a taxon and so provide important evidence of the species concept. The international codes for biological nomenclature clearly state the rules for type specimens. For example in the code for algae, fungi, and plants it states that, there should only be one holotype, lectotype or neotype, and all plant type material should be collected at the same gathering. Furthermore, even though type specimens are some of the most important specimens in a collection their associated data can be wrong, or nomenclatural decisions in the past may have been made in error. Today, there is no obvious data source where scientists can retrieve integrated information on the typification.
During this project, the goal is to explore the possibilities of enriching and increasing the visibility of type specimens through Linked Open Data (LOD) focussed on the genus Solanum.
Solanum is a large and diverse genus of flowering plants of some 800 species, which include three food crops of high economic importance: the potato, the tomato and the eggplant (aubergine, brinjal). It also contains the nightshades and horse nettles, as well as numerous plants cultivated for their ornamental flowers and fruit.

Fig. 1. A typical Solanum species, S. dulcamara.
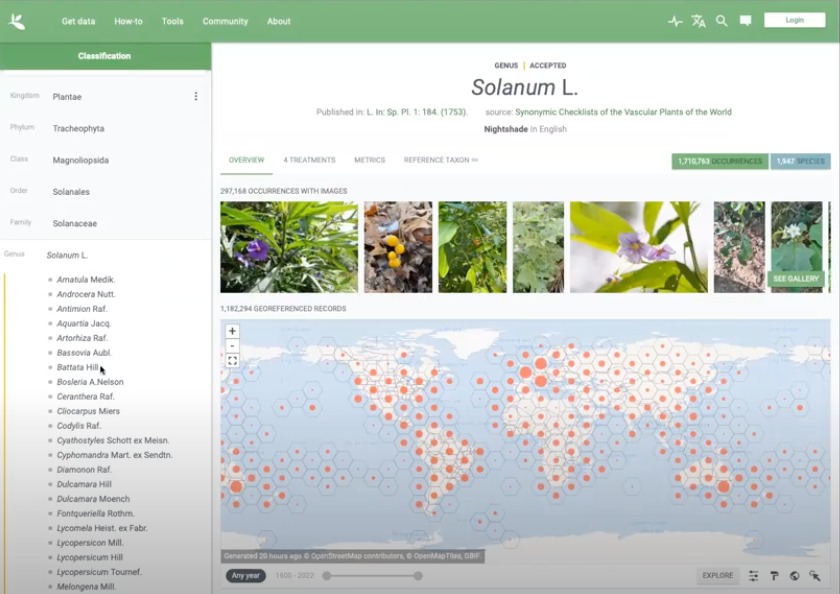
Fig. 2. Showing the cosmopolitan distribution of the genus.
Expected outcomes
The primary outcome from this project is a refined genus catalogue, including scripts to help build the catalogue, for the Solanum genus, to include not only species name but also treatments, synonyms, specimens etc.
Methods
Starting off with all types of the participating infrastructures, a methodology was developed to enrich as many types as possible with nomenclatural information as well as linking them to the treatments in which they were used. This information will then flow back to the institutional repositories and if possible be added to the machine readable data of the specimens.
Results and Discussion
A systematic approach was taken to enhance the data available for the genus Solanum. In total, in this small project, we were able to digitise 18 additional articles, liberating 184 new treatments from the literature, identifying 466 material citations and making them available for the species entry in GBIF. Using the herbarium collections, we were able to identify 1216 specimens that could be connected to their typified name. We also established or enhanced workflows that will enable this work to be done at greater scale and on different taxa groups in the future.
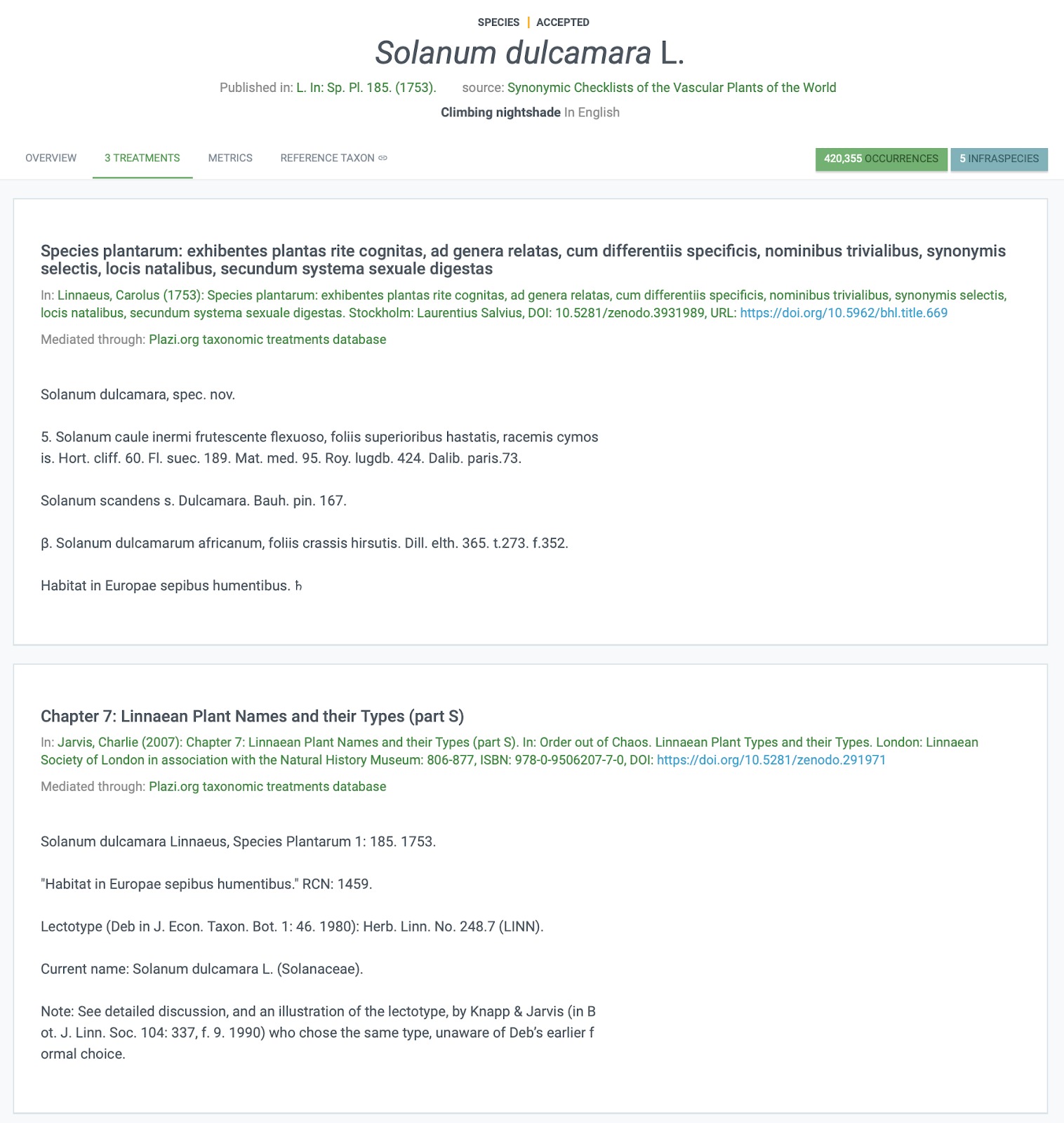
Fig. 3. Illustrating how treatment data have been added to the GBIF entry for S. dulcamara, including Linnaeus’ original description and a much later refinement of the genus from 2007.
Future Work
Solanum is a large genus, and much work remains to be done to establish additional data linkages and refinements. From a technical perspective, we can increase the visibility of the types by adding them to Wikidata. This will involve a design phase on the schema of the botanical specimens and piloting a limited number of specimens in Wikidata. If this proves to be accepted by the Wikidata community, we can add the types by writing a bot that updates available information on a regular basis.